Reproducibility of Interactive Analyses
Abstract
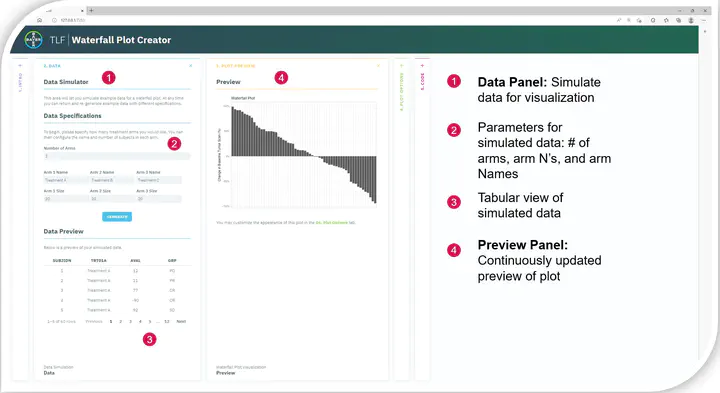
R and Shiny have seen significant adoption in pharmaceutical data science, owing to the relative ease with which an interactive application can be created and shared to non-technical audiences to engage with. They have been employed in numerous analysis and reporting activities such as delivering insight dashboards, performing exploratory analyses, generating submission dossiers, and aiding in medical review. These apps are evolving from an exploratory state to pivotal decision-making tools in many facets of clinical analysis and operations. This makes it increasingly imperative to being able to replicate the novel insights (derived from user-driven interaction) outside the application in a traceable setting that can be shared with regulators. In this paper, we share our learnings in exploring reproducibility and Shiny. We present our initial use-case, methodology and discuss both practical and technical considerations. Lastly, we outline how leveraging code reproducibility could enable efficiencies in helping programmers to learn open-source languages.